I’ve been asked to write a chapter on the role of the environment in transmission in an Springer book (on the potential role for antimicrobial surfaces in healthcare). So, I’ve been busy updating my 2011 ICHE literature review on a similar topic, drawing on an excellent recent AJIC review by Dr Donskey.
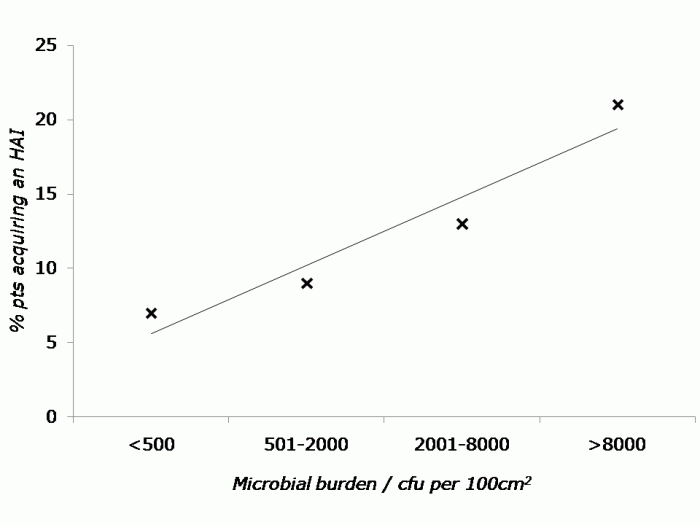
There are some epidemiological associations that suggest an important role for contaminated surfaces in transmission. Most compelling are the studies showing that admission to a room previously occupied by a patient with certain environmentally-associated pathogens increases the risk of acquisition for incoming patients, presumably due to residual contamination. However, in order to really nail a scientific association, an intervention is required. Hence, the environmental intervention studies provide the highest quality evidence evaluating the role of the environment in transmission (see the Table below).
These studies have shown that switching to more effective agents, improving the cleaning / disinfection process or turning to automated “no-touch” room disinfection systems (NTD) can reduce transmission in endemic settings. It’s important to note that some studies report an ineffective environmental intervention. These are important to publish to avoid publication bias. Looking under the bonnet of these studies usually offers an explanation as to why they did not show a significant reduction in transmission. For example:
- Wilcox 2003. There was virtually no impact on the frequency of C. difficile environmental contamination on the wards using bleach, so it’s surprising that they saw any reduction in CDI!
- Valiquette 2007. The bundle of interventions, some of which were environmental, was only given a few months to be effective.
- Wilson 2011. This one is more difficult to explain. Perhaps it was underpowered to detect a clinical impact in the declining prevalence of MRSA in the UK?
- Dharan 1999. The intervention was focused mainly on improving the cleaning and disinfection floors, which are not exactly a high-touch, high-risk sites.
Believe it or not, I still occasionally meet people who tell me that contaminated surfaces do not contribute to transmission. That rather dated viewpoint is becoming increasingly untenable as the volume and quality of data evaluating the role of the environment in transmission continues to increase. For me, the question has now moved on to how much contaminated surfaces contribute to transmission, and how best to address contamination of the hospital environment.
Table. Intervention studies evaluating the role of contaminated surfaces in the endemic transmission of nosocomial pathogens.
| Reference |
Setting, location |
Organism |
Study design |
Key findings |
| Mayfield 2000 1 |
Three units, USA |
C. difficile |
18-month before-after study of a switch from QAC to bleach disinfection. |
Significant reduction in CDI incidence on the highest risk unit from 8.6 to 3.3 cases per 1000 patient-days. |
| Wilcox 2003 2 |
Two units, UK |
C. difficile |
2-year ward cross-over study of a switch from detergent to bleach disinfection. |
Significant reduction in CDI incidence on one of the units (from 8.9 to 5.3 cases per 100 admissions), but not on the other. |
| McMullen 2007 3 |
MICU and SICU, USA |
C. difficile |
2-month before-after evaluation of bleach disinfection of CDI rooms on SICU and 4-month evaluation of bleach disinfection of all rooms on MICU in a hyper-endemic setting. |
Significant reduction in CDI incidence on both units (10.4 to 3.9 cases per 1000 patient days on SICU; 16.6 to 3.7 cases per 1000 patient days on MICU). |
| Valiquette 2007 4 |
Hospital-wide, Canada |
C. difficile |
5-month evaluation of enhanced infection control and disinfection, including a switch to bleach, and a subsequent switch to ‘accelerated’ hydrogen peroxide. |
Neither environment intervention made a significant impact on the incidence of CDI; a reduction in the use of high-risk antibiotics significantly reduced the incidence of CDI. |
| Boyce 2008 5 |
Hospital-wide, USA |
C. difficile |
20-month before-after study on the use of HPV disinfection for terminal disinfection of CDI rooms. |
Significant reduction in CDI incidence on five high incidence units (from 2.3 to 1.3 cases per 1000 patient-days). Lesser reduction in CDI incidence hospital wide. |
| Hacek 2010 6 |
Three hospitals, USA |
C. difficile |
3-year before-after study on switching from QAC to bleach for terminal disinfection of CDI rooms. |
Significant reduction in the incidence of CDI (from 0.85 to 0.45 per 1000 patient days). |
| Orenstein 2011 7 |
Two medical units, USA |
C. difficile |
2-year before-after study on switching to bleach wipes for daily and terminal disinfection of all rooms. |
Significant reduction in the incidence of CDI (from 24.2 to 3.6 per 1000 patient days). |
| Manian 2013 8 |
Hospital-wide, USA |
C. difficile |
3-year before-after study on enhanced terminal disinfection of CDI rooms using HPV and bleach. |
Significant reduction in the incidence of CDI (from 0.88 to 0.55 cases per 1000 patient days). |
| Hayden 2006 9 |
ICU, USA |
VRE |
9-month before-after study on educational improvement of cleaning and hand hygiene. |
The frequency of environmental contamination and patient acquisition of VRE were reduced from 33 to 17 acquisitions per 1000 patient-days during the improved cleaning phase. |
| Datta 2011 10 |
ICU, USA |
VRE / MRSA |
3-year before-after study of an intervention (fluorescent markers, “bucket method” and education) to enhance daily and terminal cleaning. |
Significant reduction of MRSA (3.0% to 1.5% of admissions) and VRE (3.0% to 2.2% of admissions) acquisitions; intervention significantly reduced the increased risk from the prior occupant for MRSA but not VRE. |
| Perugini 2011 11 |
Hospital-wide, Brazil |
VRE |
4-year before-after study of an educational and observational intervention for cleaners. |
Significant reduction in VRE infection (from 7.7 to 1.9 per 1000 patient days) and environmental contamination. |
| Grabsch 2012 12 |
Hospital-wide, Australia |
VRE |
18-month before-after study of a multimodal intervention (switch to bleach, improved monitoring of cleaners, modification of VRE contact isolation, periodic ‘super-clean-disinfection’ of high-risk wards). |
Significant reduction of VRE colonization (from 10.7% to 8.0% of patients) and VRE environmental contamination. |
| Passaretti 2013 13 |
ICU, USA |
VRE / all MDROs |
30-month cohort study on the impact of HPV decontamination. |
Patient admitted to rooms disinfected using HPV significantly less likely to acquire an MDRO (15.7 to 6.2 per 1000 patient days) and VRE (11.6 to 2.4 per 1000 patient days). |
| Mahamat 2007 14 |
Hospital-wide, UK |
MRSA |
8-year interrupted time series analysis of multiple infection control interventions. |
Introduction of bleach disinfection, environmental sampling, alcohol gels and admission screening all reduced the prevalence of MRSA. |
| Dancer 2009 15 |
Two wards, UK |
MRSA |
12-month cross over-study on the impact of one extra cleaner. |
Enhanced cleaning was associated with significant reductions surface contamination, hygiene fails and MRSA acquisition. |
| Wilson 2011 16 |
ICU, UK |
MRSA |
12-month randomized crossover study on the impact of additional twice daily cleaning of hand contact surfaces. |
Significant reduction in the detection of MRSA on surfaces and hands, but no significant change in MRSA acquisition was detected. |
| Dharan 1999 17 |
5 medical wards, Switzerland |
– |
4-month controlled study where 3-wards received an intervention (including an active oxygen based compound) and 2 wards continued current practice. |
Intervention associated with reduced contamination but not reduced nosocomial infection or MRSA infection / colonization. |
HPV = hydrogen peroxide vapour.
References
1. Mayfield JL, Leet T, Miller J, Mundy LM. Environmental control to reduce transmission of Clostridium difficile. Clin Infect Dis 2000; 31: 995-1000.
2. Wilcox MH, Fawley WN, Wigglesworth N, Parnell P, Verity P, Freeman J. Comparison of the effect of detergent versus hypochlorite cleaning on environmental contamination and incidence of Clostridium difficile infection. J Hosp Infect 2003; 54: 109-114.
3. McMullen KM, Zack J, Coopersmith CM, Kollef M, Dubberke E, Warren DK. Use of hypochlorite solution to decrease rates of Clostridium difficile-associated diarrhea. Infect Control Hospital Epidemiol 2007; 28: 205-207.
4. Valiquette L, Cossette B, Garant MP, Diab H, Pepin J. Impact of a reduction in the use of high-risk antibiotics on the course of an epidemic of Clostridium difficile-associated disease caused by the hypervirulent NAP1/027 strain. Clin Infect Dis 2007; 45 Suppl 2: S112-121.
5. Boyce JM, Havill NL, Otter JA et al. Impact of hydrogen peroxide vapor room decontamination on Clostridium difficile environmental contamination and transmission in a healthcare setting. Infect Control Hosp Epidemiol 2008; 29: 723-729.
6. Hacek DM, Ogle AM, Fisher A, Robicsek A, Peterson LR. Significant impact of terminal room cleaning with bleach on reducing nosocomial Clostridium difficile. Am J Infect Control 2010; 38: 350-353.
7. Orenstein R, Aronhalt KC, McManus JE, Jr., Fedraw LA. A targeted strategy to wipe out Clostridium difficile. Infect Control Hosp Epidemiol 2011; 32: 1137-1139.
8. Manian FA, Griesnauer S, Bryant A. Implementation of hospital-wide enhanced terminal cleaning of targeted patient rooms and its impact on endemic Clostridium difficile infection rates. Am J Infect Control 2013; 41: 537-541.
9. Hayden MK, Bonten MJ, Blom DW, Lyle EA, van de Vijver DA, Weinstein RA. Reduction in acquisition of vancomycin-resistant enterococcus after enforcement of routine environmental cleaning measures. Clin Infect Dis 2006; 42: 1552-1560.
10. Datta R, Platt R, Yokoe DS, Huang SS. Environmental cleaning intervention and risk of acquiring multidrug-resistant organisms from prior room occupants. Arch Intern Med 2011; 171: 491-494.
11. Perugini MR, Nomi SM, Lopes GK et al. Impact of the reduction of environmental and equipment contamination on vancomycin-resistant enterococcus rates. Infection 2011; 39: 587-593.
12. Grabsch EA, Mahony AA, Cameron DR et al. Significant reduction in vancomycin-resistant enterococcus colonization and bacteraemia after introduction of a bleach-based cleaning-disinfection programme. J Hosp Infect 2012; 82: 234-242.
13. Passaretti CL, Otter JA, Reich NG et al. An evaluation of environmental decontamination with hydrogen peroxide vapor for reducing the risk of patient acquisition of multidrug-resistant organisms. Clin Infect Dis 2013; 56: 27-35.
14. Mahamat A, MacKenzie FM, Brooker K, Monnet DL, Daures JP, Gould IM. Impact of infection control interventions and antibiotic use on hospital MRSA: a multivariate interrupted time-series analysis. Int J Antimicrob Agents 2007; 30: 169-176.
15. Dancer SJ, White LF, Lamb J, Girvan EK, Robertson C. Measuring the effect of enhanced cleaning in a UK hospital: a prospective cross-over study. BMC Med 2009; 7: 28.
16. Wilson AP, Smyth D, Moore G et al. The impact of enhanced cleaning within the intensive care unit on contamination of the near-patient environment with hospital pathogens: a randomized crossover study in critical care units in two hospitals. Crit Care Med 2011; 39: 651-658.
17. Dharan S, Mourouga P, Copin P, Bessmer G, Tschanz B, Pittet D. Routine disinfection of patients’ environmental surfaces. Myth or reality? J Hosp Infect 1999; 42: 113-117.
 Photo: Acinetobacter on MacConkey by Iqbal Osman.
Photo: Acinetobacter on MacConkey by Iqbal Osman. Photo key: from left to right: Dr. Nicholas Namias, Dr. Silvia Munoz-Price, Dr. Rossana Rosa and Dr. Daniel Kett. Location: Trauma Intensive Care Unit.
Photo key: from left to right: Dr. Nicholas Namias, Dr. Silvia Munoz-Price, Dr. Rossana Rosa and Dr. Daniel Kett. Location: Trauma Intensive Care Unit.