We often see those tasked with finding suitable isolation facilities counting down to when precaustions can be discontinued and the ’48 hours clear’ of symptoms of loose stools or vomiting has almost become one of the most welcome statements heard in healthcare settings. No more contact precautions, no more disinfectants sloshing around, normality beckons.. Or should it? Continue reading
We often see those tasked with finding suitable isolation facilities counting down to when precaustions can be discontinued and the ’48 hours clear’ of symptoms of loose stools or vomiting has almost become one of the most welcome statements heard in healthcare settings. No more contact precautions, no more disinfectants sloshing around, normality beckons.. Or should it? Continue reading
Contamination
Biofilms make the hospital environment far from ‘inanimate’
Anybody doubting that biofilms really do exist on dry hospital surfaces needs to read this study: biofilms are there, they are complex, and they are common. A landmark study by the same Australian Vickery group published in 2012 first identified biofilms on a handful of dry hospital surfaces in an ICU. But this study is far more comprehensive and convincing.
Hand Hygiene, Surfaces and Modelling
Interesting publication being highlighted as part of the WHO hand hygiene day in Leeds, UK suggests through modelling that the type of care, number of surface contacts and the distribution of surface pathogens are most likely to affect the relative quantity of pathogens accried on hands. The paper is published in ‘Indoor Air’, (not a journal that inhabits my bedside table) and we do have to remember that, as G.E.P Box stated, “Essentially, all models are wrong. But some are useful”.
What’s lurking in the NYC subway? “City-scale metagenomics” brings unprecedented resolution
A remarkable study published last week in Cell Systems has described the ‘environmentome’ of the subway system in NYC. The study has attracted a fair bit of attention in the mainstream press, not least due to claims of Bubonic Plague and Anthrax lurking in the NYC subway system (more on this later)…
The authors took a ‘microbiomic’ approach to characterize the DNA found on surfaces, similar in concept to the Hospital Microbiome Project. Since the collection of microbes in and on our bodies outnumbers our own cells by 10:1, and accounts for up to a third of active molecules in the bloodstream, this sort of approach to microbiology is going to become more and more common.
The study is mind-blowing in many ways, with 1457 samples collected from subways, public parks and alongside a canal. The headline findings are:
- DNA from pathogens (including Yesinia pestis and B. anthracis and some antibiotic resistant bacteria) were found on some surfaces.
- Human DNA identified in subway stations mirrors the demographics of the local region.
- The microbiome of surface in a station is far from stable: hourly sampling over a single day at Penn Station (a busy rail hub) identified considerable ebb and flow in predominant bacterial species.
I have a lingering concern that the techniques available for bioinformatic analysis have not yet caught up with our ability to sequence vast amounts of DNA. Put another way, the identification of bacteria in metagenomic samples is a surpassingly and rather unnervingly approximate science.
In order to identify bacteria in the sample, very many short reads of DNA are produced and then compared with databases. (This results in particular difficulty in distinguishing plasmid from genomic DNA, the subject of a recent post.) Almost half of the DNA identified in the study from New York did not match any known sequences. Furthermore, some of the species identified seem rather unlikely. For example, two of the most common Eukaryotic DNA species identified were the Mountain Pine Beetle and Mediterranean Fruit flies. Now, I’ve spent many-a-weekend in NYC from my time living in Connecticut, and I’ve never seen a Mountain Pine Beetle down there. So, this seems almost certain to be a mis-match with a closely related species due to imperfect databases. And yet when it comes to Y. pestis and B. anthracis, the authors seem more certain that the matches are correct, unlikely as they seem. (There is an acknowledgement in the discussion that these apparent “best hits” may be erroneous.)
Another key limitation is the degree of viability associated with the DNA identified. It could be that much of the DNA is a shadow of ancient contamination that is no longer viable. Whilst the authors did do a small amount of conventional sampling, and did grow some antibiotic resistant bacteria, there is no real sense of how much of the DNA identified is from viable microbes.
Quite a few years ago, I took some similar samples from buses and tube trains in London, and found no MRSA whatsoever and only a few sites grew S. aureus. It would be fascinating to see how a metagenomic analysis of these samples would look. Would there be a vastly different ‘environmentome’ in the London Underground compared with the NYC subway? Probably, but I suspect you wouldn’t find many Mountain Pine Beetles, Y. pestis or B. anthracis in either!
One of the best parts of the study is the accompanying website, which provides an interactive overview of the ‘environmentome’ of NYC: pathomap.org. Related to this, an interesting future application of these data is to derive a persons’ recent or ancient geographical location based on their current microbiome. Criminals beware – analysis of the ‘environmentome’ on the sole of your foot could invalidate your alibi!
The authors should be credited for describing the microbial ecology of the NYC subway in unprecedented detail – and this study will serve as a marker in the sand for future approaches to exploring where we fit into our inanimate environment.
Asthma and the "Hygiene Hypothesis": Does cleanliness matter? New study says “No”
Guest Blogger Prof Sally Bloomfield (bio below) writes…In proposing the hygiene hypothesis in 1989, Dr David Strachan suggested that lower incidence of early childhood infections could explain the 20th century rise in allergic diseases. This was based on data showing that larger family size appears to protect against hay fever. Strachan suggested that smaller families provided insufficient infection exposure because of less person to person spread of infections – but also because of “improved household amenities and higher standards of personal cleanliness”. From this the notion that “we have become too clean for our own good” has arisen.
Most experts now agree that the “hygiene hypothesis” is a misnomer. Although the basic concept is still seen as correct, the link to infectious disease and hygiene is now largely discounted. A number of refinements to the hypothesis offer a more plausible explanation. The Old Friends (OF) Mechanism was proposed by Graham Rook in 2003. He proposed that the required exposures are not childhood diseases such as colds, flu, measles, norovirus, which have evolved only over the last 10,000 years, but the microbes we co-evolved with in hunter-gatherer times when the human immune system was developing. These Old Friends include largely non-harmful organisms such as helminths, commensal microbiota and environmental saprophytes.
Although these microbes are still there, through lifestyle changes we have gradually lost our exposure to them. Improved water, sanitation and food quality, although protective against infections, have also inadvertently reduced exposure to Old Friends which occupy the same habitats. The decline in natural childbirth in favour of caesarean section, and bottle instead of breast feeding is another likely factor. Reduced exposure to our outdoor environment has also occurred – we now spend up to 80% of our time indoors. Also, antibiotics may alter our interaction with microbes leading to reduced diversity of human gut microbiota.
Despite a shift in scientific thinking, the so-called “hygiene hypothesis” is still widely accepted in the public domain and still discussed in terms of concepts such as ‘eat dirt’, ‘too clean is bad’, or ‘sterile homes’. The lay audience, and even the U.S. FDA attribute the problem to ‘the extremely clean household environments often found in the developed world’.” (See headlines below!)
What is overlooked is the fact that the relationship between household or personal cleanliness and development of allergies has never been properly investigated. At last – just this week, we have seen publication of the first study to directly evaluate this issue. From the study, Erika von Mutius, a highly respected researcher in this field, concludes – No. “Development of allergies and asthma was not related to cleaning activities”.
Methods: The study involved a birth cohort of 399 participant families recruited in urban and suburban regions of Munich, Germany, between Oct 1999 and Dec 2001. A telephone questionnaire interview comprising 31 questions was carried out to assess cleaning habits and cleaning frequencies in the homes, the use of detergents and the personal cleanliness of the child. In addition, 13 lifestyle factors and home characteristics were obtained from a self-administered questionnaire. Questions about the child’s health focused on respiratory and allergic problems. Bacterial markers of home cleanliness were assessed in samples of floor and mattress dust.
Results and conclusions: As found by other workers, bacterial exposure in house dust was found to be associated with reduced risk of childhood allergies. In turn, personal cleanliness, such as washing hands, and home cleanliness were objectively reflected by dust parameters. However, neither personal nor home cleanliness were associated with protection from asthma and allergies (see flow chart, below).
Note: In this study, cleanliness was substantiated by rather unspecific dust measurements. The findings however suggest that allergy protection operates through as yet unknown exposures, not assessed by unspecific markers. Future studies will require more detailed microbial analysis. Whether these microbes are affected by cleaning remains to be elucidated, but findings with unspecific markers suggest that normal cleaning does not affect permanent microbial colonization of indoor environments.
If, as now seems, allergies are not the price we have to pay for protection from infection, two fundamental questions need to be addressed:
- “How can we develop an approach to hygiene, which helps to reconnect us with the necessary microbial exposures, whilst also protecting us against infectious diseases? “
- How do we change public understanding about the difference between “cleanliness” (absence of visible dirt) and “hygiene” (protecting against infectious diseases)?
The answer to the first question is a return to basics, which means promoting “targeted hygiene”. This means identifying the critical points in the chain of infection transmission and applying effective hygiene procedures at the appropriate times to prevent further spread. Appropriate times are those associated with activities such as food and water hygiene, respiratory hygiene, toilet hygiene, laundry hygiene and so on.
Dispelling the misconceptions is a real challenge, made more difficult by the fact that people tend to think that hygiene and cleanliness is the same thing (i.e “if it looks clean it must be germ free”). It is possible that this is best done by promoting a more constructive approach i.e . stressing that getting dirty is healthy, but hygiene is vital in the times and places that matter.
Further Reading: Bloomfield SF, Stanwell-Smith R, Rook GA. 2013. The hygiene hypothesis and its implications for home hygiene, lifestyle and public health: summary.
The study can be found at: Am J Respir Crit Care Med. 2015 Jan 13. [Epub ahead of print] Asthma and the Hygiene Hypothesis – Does Cleanliness Matter? Weber J, Illi S, Nowak D, Schierl R, Holst O, von Mutius E, Ege MJ.
Guest blogger bio:
Dr Sally Bloomfield is an Honorary Professor at the London School of Hygiene and Tropical Medicine. She is also is the Chairman and Member of the Scientific Advisory Board of the International Scientific Forum on Home Hygiene (IFH). Through these roles Professor Bloomfield continues to develop her work in raising awareness of the importance of home hygiene in preventing the transmission of infectious disease, and developing and promoting home hygiene practice based on sound scientific principles. She is also working to develop understanding of “hygiene issues” such as the “hygiene hypothesis” and “antimicrobial resistance”.
Professor Bloomfield’s background is in healthcare and infectious disease. She has a degree in Pharmacy, and PhD in Pharmaceutical Microbiology from the University of Nottingham. Sally was previously a Senior Lecturer in Pharmaceutical Microbiology at Kings College London (1995 – 1997) and a Hygiene Liaison manager at Unilever Research Port Sunlight UK (1997 – 2001). She has published 100 research and review papers on the subject of home hygiene and the action and mode of action role of antimicrobial agents.
CRE “trafficking” plasmids through hospital surfaces
A team from the NIH Clinical Center in the US present a fascinating study, exploring the transmission of carbapenemase-encoding plasmids in unprecedented detail. The intro does a good job of introducing the ‘triple threat’ from CRE: pan-drug resistance, sharply increasing prevalence, and the potential for the horizontal transfer of carbapenemase genes between Enterobacteriaceae species. They introduce the idea of “plasmid trafficking”, which evokes images of shady bacteria dealing in antibiotic resistance genes (a la the infamous cartoon below):
NIH is a hospital that takes CRE seriously, after being stung by an outbreak in 2011. A quick look at who they screen for CRE illustrates just how seriously they take the threat:
- ICU / high-risk patients screened twice weekly.
- All patients screened monthly.
- Admissions from other hospitals screened for CRE…twice (and given pre-emptive contact precautions until negative cultures are confirmed, for good measure).
They also performed some environmental sampling and recovered several CRE from the hospital environment. This will surprise some, but Enterobacteriaceae do have the potential to survive on surfaces for longer than you may expect.
Surveillance cultures identified 10 patients with KPC-producing Enterobacteriaceae and environmental surveillance identified 6 KPC-producing Enterobacteriaceae. They combined these with several historic isolates from the 2011 outbreak, and a couple of imported isolates to give a sample size of 20 isolates. They wanted to dig deeper into these isolates to explore whether or not they shared any plasmids. And here’s where it gets rather complicated. Conventional whole genome sequencing produces many short reads (100-500 bp) but these cannot distinguish between plasmids and chromosome-encoded genes. Therefore, the authors used a technique called single-molecule, real-time (SMRT) to generate longer reads (around 1000 bp) that make it possible to distinguish between plasmids and chromosome-encoded genes. [I know that I’ve over-simplified this clever genomics massively – but I’ll quickly get out of my depth otherwise!]
The report presents a picture of rare patient-to-patient nosocomial transmission (only 1 of 10 patients were thought to be in-hospital acquisitions), continual importation of diverse CRE, and a complex network of even more diverse plasmids. To illustrate the diversity, one strain of CRE contained no fewer than three distinct KPC-encoding plasmids!
The authors find some evidence of environmental spread of carbapenemase-encoding plasmids, with the carbapenemase-encoding plasmid from a patient matching plasmids recovered from different species of Enterobacteriaceae found in the patient’s environment. What the authors did not demonstrate is transmission of carbapenemase-encoding plasmids from the environment to patients – but I wouldn’t want to be admitted to a room with CRE lurking in the hospital environment!
There’s quite a bit of science around the horizontal transmission of plasmids within biofilms. Combine this with the recent finding of biofilms on dry hospital surfaces, and you have a concerning new angle on how CRE may be transmitted in hospitals.
Image credit. Nick Kim, with permission.
Article citation: Conlan S, Thomas PJ, Deming C et al. Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci Transl Med 2014; 6: 254ra126.
Is deliberately seeding hospital rooms with Bacillus spores a good idea? No, I don’t think so either!
A fascinating Italian/Belgian multicentre study introduces us to the idea of “biocontrol” for problematic surface contamination. They test using “live” cleaning products that deliberately seed hospital surfaces with Bacillus species spores in an attempt to reduce the ecological space for pathogenic microbes through a “competitive exclusion” approach. Ridiculous as it sounds, there’s some logic to this idea. We’re just beginning to understand the potential of complementing a depleted microbiome in human health, so perhaps the same theory goes for the “environmentome”?
The study design is on the one hand impressive and ambitious, with more than 20,000 surfaces samples collected from the three hospitals. However, it is also messy and confusing, with different intervention and sampling protocols in the three hospitals. In particular, it’s a real shame that areas were not randomized to receive the “live” vs. conventional cleaning agents. It seems clear that this was not a carefully planned multicentre study using a standardized protocol – it reads more like three separate studies shoe-horned together.
That said, the results are impressive. Areas treated with the “live” cleaning agents were significantly less likely to be contaminated with coliforms, S. aureus, Candida albicans, with a more moderate impact on C. difficile. However, it’s difficult to determine the scale of the reduction since the relative rather than actual load reductions are reported.
A neat sub-experiment at one of the hospitals is perhaps the most convincing part of the study, where conventional and “live” cleaning agents were alternated (Figure). You can clearly see that the microbial load tracked downwards when the “live” agent was used, and rebounded when the conventional agent was reinstated.
 Figure: Bacterial load of coliforms (black circles) and S. aureus (white circles). Black arrow = beginning of the “live” cleaning agent; black dotted arrow = conventional cleaning agent.
Figure: Bacterial load of coliforms (black circles) and S. aureus (white circles). Black arrow = beginning of the “live” cleaning agent; black dotted arrow = conventional cleaning agent.
Notwithstanding the impressive reductions, this approach is ringing some alarm bells:
- Do we really know what we’re doing by deliberately seeding the hospital environment with bacterial spores? Almost all microbes can be pathogenic to immuno-compromised patients. Plus, whilst you know what you’re putting down, you don’t know what it will become when exposed to the selective pressure of hospitals. The authors did take a look at this, using antibiotic susceptibility testing and a PCR assay to show that Bacillus species identified from the original cleaning agents and from hospitals surfaces during study did not differ in their carriage of antibiotic resistance genes. However, this is only scratching the surface of a complex risk.
- Where do all the pathogens go? Having an environment that is full of Bacillus spores does not make a scrap of difference to the amount of pathogens that are shed into the environment. So, either the Bacillus spores somehow reduce the amount of time that these pathogens survive on surfaces, or offer them a more complex hiding place. I suspect the latter is more likely.
- Related to this, recent work has identified established biofilms on dry hospital surfaces with important implications. Won’t a daily dose of Bacillus spores only serve to promote the buildup of this biofilm?
- The authors proffer some potential reasons for the lower bacterial counts, including competition for nutrients and quorum sensing to destabilize biofilms. I think these are very unlikely, because they rely on the Bacillus spores germinating on the surfaces. I suspect that the spores remain firmly as spores, and the reductions are explained by occlusion and competition for space.
- Ethics can be a pain, but it’s there for a reason – to prevent our patients from unnecessary harm. The outcome of their ethical submission was surprising: “The two Ethics Committees stated that a formal authorization was not necessary because the probiotic products would not be directly administered to patients but exploited for cleaning of hospital surfaces only.” Applying a soup of Bacillus species spores to a patient’s room is pretty much the same thing as applying the soup directly to their skin. Personally, I’d like to choose whether or not I’m admitted to a room deliberately seeded with Bacillus spores!
- The authors insist on calling the “live” cleaning agents ‘probiotics’, which seems misplaced. To me, ‘xxx-biotics’ implies something that is administered to a patient.
The use of “live” cleaning agents provides an interesting alternative approach to antimicrobial surfaces, or chemicals with residual biocidal activity. However, I am not sure I accept the authors stark choice as their final conclusion: ‘When it comes down to risk management, one has to decide whether a patient should stay in an environment dominated by food grade microorganisms or in an environment harboring an elevated level of increasingly resistant pathogens.’ Personally, I’d prefer to be cared for in an environment with minimal levels of bacterial contamination, and free from contamination with pathogens. Is that too much to ask?
Article citation: Vandini et al. Hard Surface Biocontrol in Hospitals Using Microbial-Based Cleaning Products. PLoS One 2014;9:e108598.
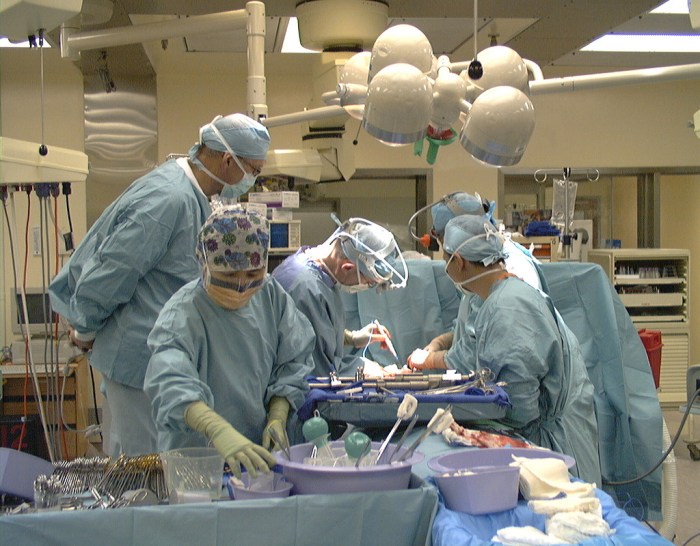
The inanimate environment doesn’t contribute to pathogen transmission in the operating room…OR does it?
Nosocomial or hospital-acquired infections are a worldwide problem affecting millions of patients yearly and increasing morbidity and mortality. The role of the hospital inanimate environment (environmental surfaces and surfaces of medical equipment) in the transmission of certain nosocomial pathogens such as C. difficile, norovirus, MRSA, VRE and Acinetobacter is now well established supported by various studies and publications. Most, if not all of these studies, investigated the transmission process in patient rooms or ICUs. Although the role of air in the transmission of pathogens has been extensively studies in the operating room (OR) setting, do contaminated surfaces play a role in pathogen transmission in the OR?
A recent review article published in the journal “Surgical Infection” questioned whether the OR inanimate environment contributed to the transmission of pathogens, hence possibly causing infections including surgical site infections (SSIs). Few studies have investigated surface contamination in the OR and even fewer have investigated possible pathogen transmission from the environment in this setting. While the inanimate environment in the OR has been considered a potential source for pathogens that may cause SSIs for more than 100 years, the role of this environment in the patient acquisition process within this setting is still debatable. Before revealing the conclusions of the review paper, I would like to look at both sides of the argument.
THE OR INANIMATE ENVIRONMENT DOES NOT PLAY A ROLE IN PATHOGEN TRANSMISSION AND INFECTION
The patient population and length of stay
In a hospital, patients colonised or infected may spend days or even months in ward rooms or ICUs increasing the chance that these patients will contaminate their environment or acquire pathogens from that environment. The likelihood of environmental contamination or pathogen acquisition increases with the length of hospital stay as well as other factors such as gross contamination and soiling.
In the OR setting however most patients spend only few hours under full or partial anaesthesia. This makes it less likely that these patients will contaminate their environment or acquire pathogens from the environment (by self inoculation at least). In addition, although gross contamination via blood for example is common, other type of gross environmental contamination linked to transmission such as diarrhoea and vomiting are less likely to occur in an OR.
The OR environment (surfaces and air)
Unlike most patient rooms, OR air quality is well regulated to prevent contamination via the air. This not only reduces the risk of infection via airborne pathogens but also reduces the amount of pathogens settling on and contaminating environmental surfaces in ORs. In addition, the OR inanimate environment is routinely cleaned/disinfected. Most ORs are cleaned at the end of the working day and many surfaces and areas are cleaned before and between surgeries with strict policies on how to deal with gross contamination (e.g. blood and tissue).
Minimising infection risk
As most SSIs are thought to originate from patients’ or healthcare personnel’s own flora, many interventions are in place in ORs to minimise the risk of contamination and infection. These include policies for hand scrubbing and disinfection, gloving, masks, and the proper preparation of patients’ skin before incision. The instruments used in surgery are also routinely sterilised before surgery to minimise the risk of infection.
Organisms involved in SSIs
The hospital environment has been implicated in the transmission of a number of pathogens including norovirus, C. difficile, MRSA, VRE and Acinetobacter. These pathogens are able to contaminate the environment at a high load and survive for long period of time facilitating transmission and acquisition. While infections with these organisms can be acquired in the OR, with the exception of Staphylococcus species, these pathogens are not the major causes of SSIs. The environmental resilience of other organisms involved in SSIs is not well characterised and it is unclear whether they can survive long enough in the environment to be transmitted.
THE OR INANIMATE ENVIRONMENT IS A SOURCE OF PATHOGENS THAT CAUSE INFECTION
The OR environment
ORs are busy, with many personnel involved during a surgical procedure, some of whom come and go in and out of the OR during the process. It is also an environment with multiple and frequent contact between personnel, patients and the environment including medical equipment. It is difficult if not impossible to observe the WHO’s 5 moments for hand hygiene in such an environment, or to clean and disinfect the environmental surfaces effectively during a surgical procedure. Organisms originating from the floor of the OR can also be disturbed by walking and are taken into the air which may increase the risk of infection.
The OR inanimate environment is contaminated
Many people in the general public think of ORs as ultra clean, even sterile, environments. For anyone working in ORs, it is clear that this view is far from the truth. Although modern ORs have strict measures to reduce contamination, the OR inanimate environment becomes contaminated with various organisms including those involved in SSIs. Studies have reported contamination of various OR areas such as anaesthesia equipment, beds, intravenous pumps and poles, computer keyboards, telephones and OR floors. A variety of pathogens capable of causing infections have been identified including Gram-negative bacilli such as Acinetobacter and Pseudomonas species, Staphylococcus including (MRSA) and Enterococcus. These results may be in part due to the fact that suboptimal cleaning in ORs is a widespread issue in hospitals.
Pathogen transmission occurs in ORs
A number of studies in ORs focusing on the role of anaesthesia equipment and providers in the contamination and transmission of pathogens in ORs have concluded that the hands of anaesthesia providers, patient IV tubing and the immediate patient environment were contaminated immediately before or during patient care with a wide range of bacterial pathogens leading to transmission. Transmission of pathogens from and to the hands of the anaesthesia providers involving the inanimate environment occurs frequently given the frequent contact with the environment in ORs.
Human behaviour in ORs contributes to environmental contamination and transmission
We are all familiar with the view that surgeons tend to be the worst healthcare workers as far as hand hygiene compliance is concerned. However, this is only the tip of the iceberg regarding lapses in infection prevention in ORs. For instance, anaesthesia provider’s behaviour and attitude including confusion on when and how often to perform hand hygiene during a procedure is a common cause of pathogen transmission. In one study, anaesthesia providers touched 1,132 objects during 8 hours of observations in OR, but only performed a total of 13 hand disinfections. No hand disinfections were witnessed at any time during 3 (43%) of the procedures observed. Furthermore, hand hygiene failed to precede or follow procedures, blood exposure or contact with the floor. Alarmingly, it has been reported that objects that fall onto the OR floors during surgery were frequently placed back either on to horizontal work surfaces or even on to the patients themselves during operations.
THE CONCLUSION
It is clear that the inanimate environment of the OR, including medical equipment, can become contaminated with pathogens that cause infections including SSIs. These pathogens can then be transmitted to the hands of healthcare workers and have the potential to cause infection. Further studies are necessary to quantify the role of contaminated surfaces in the transmission of pathogens and to inform the most effective environmental interventions in the ORs. Given the serious consequences of SSIs, special attention should be given to the proper cleaning and disinfection of the inanimate environment in ORs in addition to the other established measures to reduce the burden of SSIs. These include addressing the human behaviour that contributes to environmental contamination and transport of surface pathogens into the vulnerable sites of patients during surgery. Such measures include reducing human traffic in ORs, stricter adherence to the standard operating protocols during procedures, and compliance with proper hand hygiene and gloving. Specific hand hygiene guidelines tailored to OR personnel may be needed given the large number of hand contact events per hour in these settings.
Image: NIH Library.
Acinetobacter contamination: is anywhere safe?
A study from New York City describes an environmental survey of contamination with antibiotic-resistant Gram-negative bacteria on surfaces in the community. The authors hypothesise that resistant Gram-negatives could be carried by staff, patients and visitors beyond the confines of the hospital.
Almost 500 environemntal samples were collected from surfaces in the public areas of six hospitals and surrounding communities (<1 mile from the hospital) (443 samples), with a further surfaces from communities >1.5 miles from any hospital as a control (50 samples). A total of 70 GNR were identified (Figure), mostly fairly inoccousous species from a human disease viewpoint. However, some potential human pathogens were identified (Table).
Figure: breakdown of Gram-negative rods identified from surfaces in public areas of the hospital and surrounding community.
Table: potential human pathogens identified from surfaces in public areas of the hospital and surrounding community.
| n | % | Species |
| 15 | 3.0 | Acinetobacter baumannii |
| 3 | 0.6 | Citrobacter freundii |
| 2 | 0.4 | Escherichia coli |
| 2 | 0.4 | Stenotrophomonas maltophilia |
| 1 | 0.2 | Enterobacter cloacae |
Some other important findings:
- All of the A. baumannii isolates were resistant to ceftazidime, and one was resistant to imipenem (i.e. carbapenem-resistant). Eleven of the 15 were clonally related to one another and to a patient isolate from one of the hospitals.
- One of the S. maltophilia isolates carried an integron-encoded VIM carbapenemase, which is potentially transmissible to other Gram-negative species (including Enterobacteriaceae).
- Each sample was cultured in an enrichment broth, and the broth was probed for the presence of a range of beta-lactamase genes (including ESBLs and carbapenemases). No beta-lactamases were detected (other than the S. maltophilia isolate). I suspect the picture would have been rather difference in New Dehli!
- Although the survey included both surfaces in public areas of hospitals and in the community, it seems that most of the A. baumannii were identified on surfaces in the community.
So, is it a surprise to see environmental contamination with antibiotic-resistant Gram-negatvies on touch-surfaces in the community? Not really, A. baumannii in particular can survive on surfaces for ages, and ‘mimics’ Gram-positive bacteria in terms of its environmental longevity (i.e. months / years). That said, I performed a similar study looking for MRSA on touch surfaces in the community in London, and didn’t find any. More importantly, do we need to do anything about this? As the authors state, A. baumannii can be virtually impossible to eliminate from hospital surfaces without resorting to hydrogen peroxide vapour. So is it time to roll hydrogen peroxide vapour into your local Pizza Hut? Clearly not. You’d hope that cleaning and disinfection protocols, which should deal with this sort of contamination, are already established in these public places, but it would be prudent to reinforce these basic hygienic practices. Also, I agree with the authors that these findings represent and opportunity for the promotion of hand hygiene in the community.
The authors use strong words to describe NYC as ‘plagued’ with resistant Gram-negative bacteria, and a ‘dismal failure to control A. baumannii.’ If this epidemic continues, we can expect to see the focus of the problem – and the target for our interventions – shift from the acute hospital setting to encompass the community.
Are contaminated hands more important than contaminated surfaces?
Cast your minds back to the 2010 HIS conference in Liverpool and Drs Stephanie Dancer and Stephan Harbarth debating the relative importance of contaminated hands vs. surfaces in the transmission of MDROs. I don’t remember the details of the debate, but I do remember the surprising lack of evidence on both sides. Back then, we had no real way to quantify the contribution of the environment to the transmission of MDROs, or to measure the relative importance of contaminated hands vs surfaces. The evidence has evolved to the extent that a group of US researchers have published a paper modeling the relative contribution of contaminated hands vs surfaces to the transmission of MDROs. I like the paper very much, and the authors should be congratulated for breaking new ground in understanding transmission routes of MDROs.
The model simulates patient-to-patient transmission in a 20-bed ICU. The values of the parameters that were used to build the model were sensible on the whole, although baseline hand hygiene compliance was set at 57-85% (depending on staff type and whether at room entry or exit), which seems rather generous when baseline environmental cleaning compliance was set at 40%. Also, the increased risk from the prior room occupant for MRSA and VRE was set at 1.4 (odds ratio) for both, whereas it probably should be higher for VRE (at least >2) based on a number of studies.
100 simulations were run for each pathogen, evaluating the impact of step-wise changes in hand hygiene or terminal cleaning compliance. The key finding is that improvements in hand hygiene compliance are more or less twice as effective in preventing the transmission of MDR A. baumannii, MRSA or VRE, i.e. a 20% improvement in terminal cleaning is required to ‘match’ a 10% improvement in hand hygiene compliance. Also, the relationship between improved terminal cleaning and transmission is more or less linear, whereas the relationship with hand hygiene shows relatively more impact from lower levels of hand hygiene compliance (see Figure, below). Thus, the line for improving hand hygiene or terminal cleaning would intercept and indeed cross over at around 40 or 50% improvement. The implication here is that hand hygiene is more important at low levels of compliance, whereas terminal cleaning is more important at high levels of compliance (although don’t forget the difference in the baseline compliance ‘setpoint’.
 Figure. The impact of percentage improvement in hand hygiene or terminal cleaning on the transmission of MDROs. Dotted line represents my not-very-scientific extrapolation from eyeballing the data.
Figure. The impact of percentage improvement in hand hygiene or terminal cleaning on the transmission of MDROs. Dotted line represents my not-very-scientific extrapolation from eyeballing the data.
The study raises some important issues for discussion:
- It had not struck me before that the level of compliance with hand hygiene and environmental cleaning are nearly identical, on average, with only around 40% of hand hygiene opportunities met and 40% of environmental surfaces cleaned if human beings are left to their own devices. Both of these figures can be improved considerably with concerted effort, but the sustainability of these improvements without continued effort is rather disappointing.
- The models address MRSA, VRE and MDR A. baumannii transmission. It’s a little strange that C. difficile was not included, since most consider this to be the ‘most environmental’ hospital pathogen.
- The study only modeled the impact of terminal cleaning, whereas daily cleaning seems likely to also be an important factor (which is acknowledged as a limitation in the discussion). This seems especially important in light of data that touching a contaminated surface carries approximately the same risk of hand contamination as touching an infected or colonized patient.
- I am not certain that this assumption makes logical sense: ‘thoroughness of cleaning of 40% implies that, given a single cleaning opportunity, there is a 40% probability that the room will be cleaned sufficiently well to remove all additional risk for the next admitted patient’. This would be true if cleaning was performed to perfection 4 times out of 10, whereas it is actually performed with 40% efficacy 10 times out of ten! To this end, it would be interesting to insert the various automated room disinfection systems into the model to evaluate and compare their impact. Indeed, hydrogen peroxide vapour has been shown to mitigate and perhaps even reverse the increased risk from the prior room occupant (for VRE at least).
- In the introduction, the authors comment that ‘A randomized trial comparing improvements in hand hygiene and environmental cleaning would be unethical and infeasible.’ I see what they’re saying here, in that it would be unethical by modern standards to investigate the impact of no hand hygiene or no environmental cleaning (although this has been done for hand hygiene), but it would be useful, feasible and ethical to perform a cluster RCT of improving hand hygiene and environmental cleaning. It would look something like the classic Hayden et al VRE study, but with an RCT design.
- How useful is mathematical modeling in informing decisions about infection prevention and control practices? This is not the first mathematical model to consider the role of the environment. For example, researchers have used models to evaluate the relative importance of various transmission routes including fomites for influenza. But a model is only as good as the accuracy of its parameters.
- Does this study help us to decide whether to invest in increasing hand hygiene or terminal cleaning? To an extent yes. If you have awful compliance with both hand hygiene and terminal cleaning at your facility, this study suggests that improving hand hygiene compliance will yield more improvement than improving terminal cleaning (for A. baumannii, MRSA and VRE at least). However, if you have high levels of compliance with hand hygiene and terminal cleaning, then improving terminal cleaning will yield more.
In general, this study adds more evidence to the status quo that hand hygiene is the single most effective intervention in preventing the transmission of HCAI. However, in a sense, the hands of healthcare workers can be seen as high mobile surfaces that are often contaminated with MDROs and rarely disinfected when they should be!